![[Logo CIB]](/imagenes/logo_cib.jpg) CENTRO DE INVESTIGACIONES BIOLÓGICAS
CENTRO DE INVESTIGACIONES BIOLÓGICASDepartment of Cell and Developmental Biology
Molecular Biology of Chromosomes
Most relevant achievements
![[Logo CIB]](/imagenes/logo_cib.jpg) CENTRO DE INVESTIGACIONES BIOLÓGICAS
CENTRO DE INVESTIGACIONES BIOLÓGICAS
Department of Cell and Developmental Biology
Molecular Biology of Chromosomes
Most relevant achievements
![]()
Most relevant achievements
In our group we have been studying the topology and replication of DNA in different prokaryotic and eukaryotic systems for the last 20 years. We were the first to characterize unidirectional DNA replication by means of two-dimensional agarose gel electrophoresis (Martín-Parras et al., 1991). We were the first to report that in those plasmids and minichromosomes containing more than one potential origin, initiation of DNA replication occurs at only one of the potential origins per replication round, regardless of origin orientation (Schvartzman et al., 1990; Martín-Parras et al., 1992; Viguera et al., 1996). This phenomenon is called "origin interference" and its nature is still unknown. In those bacterial plasmids with inversely oriented unidirectional origins, the silent origin acts as a polar barrier for the replication fork initiated at the other origin (Viguera et al., 1996). This is due to the incompetence of Escherichia coli DnaB helicase, which leads the replication fork in vivo, to unwind the RNA 3' end of RNA-DNA hybrids (Santamaría et al., 1998). Blockage of the replication fork leads to the accumulation of replication intermediates (RIs) containing an internal bubble, which may serve as substrate for inadvertent knotting events resulting from topo II-type action that tend to fix some of the crossings present in interwound supercoiled molecules (Olavarrieta et al., 2002a,b,c; Viguera et al., 1996). For this reason most of these nodes have a positive sign (Sogo et al., 1999). Blockage of the replication fork is not just a pause but permanent, as it leads to a premature termination event (Santamaría et al., 2000). Interestingly, RIs containing an internal bubble may undergo reversion of the fork, at least in vitro (Viguera et al., 2000). We were also among the first to characterize positive-supercoiling induced replication-fork reversal (Olavarrieta et al., 2002; Viguera et al., 2000; Fierro-Fernández et al., 2007a,b). In eukaryotic plasmids with a bidirectional origin, termination does not occur at a fixed site but may take place anywhere depending on the firing synchrony of both forks at the origin and the rate of forks progression (Santamaría et al., 2000). We were also the first to use two-dimensional agarose gel electrophoresis and electron microscopy to identify all the different topological forms that autonomously replicating circular plasmids and minichromosomes can adopt in living cells (Martín-Parras et al., 1998; Mayán-Santos et al., 2007; Schvartzman et al., 2010). Our group participated also in a joint effort to find that the genome of Streptococcus pneumoniae is organized in topology-reacting gene clusters (Ferrándiz et al., 2010). We also found that in Saccharomyces cerevisiae, those circular minichromosomes containing the rDNA replication fork barrier (RFB) are unstable in ?sir2 strains and integrate at the rDNA locus by homologous recombination (Benguría et al., 2003). Curiously, integration occurs only at two specific sites: the RFB and the 3’ end of the 35S precursor gene (Mayán-Santos et al., 2008). We were the first to characterize the interplay of supercoiling and decatenation during the segregation of sister duplexes in bacterial plasmids (Martínez-Robles et al., 2009). Finally, we demonstrated that positive supercoiling drives decatenation by topoisomerase II in eukaryotes (Baxter et al., 2011).
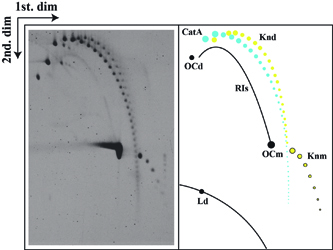 |
Autoradiogram of a 2D gel corresponding to pBR18 isolated from DH5alphaF' E. coli cells after exposure to norfloxacin and complete digestion with the single-stranded DNA nicking enzyme Nb-BsmI. A diagrammatic interpretation of the autoradiogram is shown to the right where CatAs are depicted in blue. RIs=nicked replication intermediates; Knd=nicked knotted dimers; OCd=open circles corresponding to dimers; OCm=open circles corresponding to monomers; Knm=nicked knotted monomers; Ld=linearized dimers; Lm=linearized monomers. (Martínez-Robles et al., 2009). |
 |
Cartoon illustrating changes in the topology of a DNA replication intermediate (RI) caused by fork reversal.The effect of 0.1 M NaCl at 65°C on a nicked RI is shown step by step. (A) Partially replicated RI showing a nick in the unreplicated portion. (B) Regression of one of the forks leads to the formation of a HLJ. (C) As regression of the fork continues the nascent–nascent duplex lengthens, leading to a corresponding reduction in the size of the replication bubble. (D) Total extrusion of the nascent–nascent duplex leads to two independent molecules: a nicked circular form identical to an open circle (OC) and a linear form. The parental duplexes are indicated in blue and green, and the nascent strands are depicted in red (Fierro-Fernández et al., 2007). |
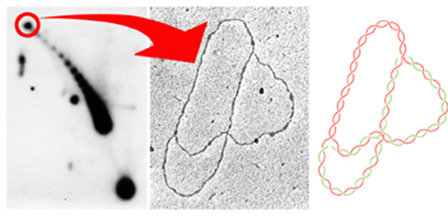 |
To
study the topology of partially replicated molecules, we constructed
a series of pBR322 derivatives where the TerE replication terminator
was cloned at variable distances from the ColE1 origin. The autoradiogram
to the left corresponds to the intact forms of the plasmid pBR18-TerE@EcoRI
analized by 2D agarose gel electrophoresis. The signal corresponding
to the relaxed forms of this partially replicated molecule is
encircled in red. These molecules were isolated from 2D gels
and examined at the electron microscope as indicated by the arrow.
The middle image corresponds to an electron micrograph of one
of these molecules. The diagram to the right represents the photographed
molecule where the parental DNA strands appear in red while the
nascent strands appear in green. As expected the unreplicated
arm corresponds to 43% of the plasmid whereas each one of the
daughter arms corresponds to 57% of the plasmid (Olavarrieta
et al., 2002). |
Identification of Knotted Replication Bubbles
 |
The
analysis of partially replicated molecules by 2D agarose gel electrophoresis
and electron microscopy allowed us to show that knots can occur between
the sister duplexes in the replicated region behind the fork. Most of the
nodes of these knotted replication bubbles have a positive sign. |
Positive supercoiling
leads to replication fork reversal
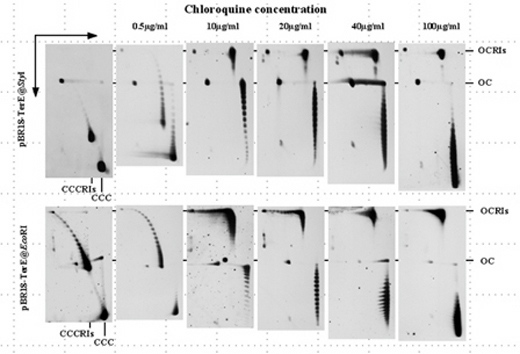
The
analysis of intact forms of plasmids containing a stalled fork (pBR18-TerE@StyI
y pBR18-TerE@EcoRI) by 2D gels allowed us to show that contrary
to what happens in the case of non-replicating plasmids (CCC),
chloroquine is unable to induce the positive supercoiling of
partially replicated molecules (CCCRIs). This occurs because
in these partially replicated intermediates positive supercoiling
is absorbed by replication fork reversal with the concomitant
formation of Holliday-like junctions (Olavarrieta
et al., 2002). |
 |
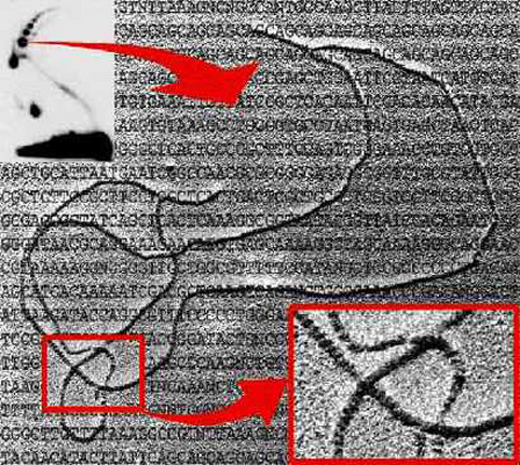 |
The
sample (pHH5.8) was analyzed by two-dimensional (2D) agarose gel electrophoresis
after digestion with AlwNI (insert at the upper left corner), enriched
for the intermediates indicated by the base of the arrow, the DNA coated
with recA and examined under the electron microscope (Sogo
et al., 1999). The insert at the lower right corner corresponds
to a higher magnification where it can be clearly noticed which
segment goes above and which below at each individual crossing.
(Electron microscopy: José Manuel Sogo, Zurich, Swizerland). |
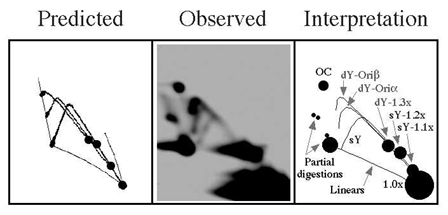
One of the techniques we use
in the lab is "Two-Dimensional (2D) Agarose Gel Electrophoresis".
Bonita Brewer and Walton Fangman (from Seattle, Washington, USA)
developed this technique in 1987 to find out the shape of the
replication intermediates (RIs) for any given linear DNA molecule.
To assist scientists to take full advantage of this technique,
we developed a computer program that predict the 2D gel patterns
expected for the RIs of any given linear DNA fragment when analyzed
by neutral/neutral 2D gels (Viguera
et al., 1998). To download our 2D gel program click
here. |
 |
| Author: Jorge Bernardo
Schvartzman Blinder Madrid, November 2003 |
Docmaster: Sonia
Centeno Madrid, 29 September 1999 |